Why Reproducible Neuroimaging
Neuroimaging, like many fields of science, has been described as having a “reproducibility crisis”. We most often hear about this crisis in the context of trust in science and the ability to reproduce the results of studies published by others. It is, of course, frustrating when you waste time and resources on trying to replicate others’ research only to find that the experiment failed. We know that some of the causes of the current reproducibility crisis are poor study design, including underpowered studies. But it is also true that even in a properly designed neuroimaging study, more and more evidence has emerged that any set of results is very sensitive to even small variations in computational environments, preprocessing steps, statistical models and analyses used. In fact, as we state in our 2019 paper, when it comes to reproducibility in neuroimaging, “Everything matters”.
Reproducible neuroimaging is therefore not only a concern to those outside your lab trying to build on your work, but to you as a researcher, project director or principal investigator. Can you reproduce your own research? Can your graduate students? Can your present and future post-docs? If a reviewer asks you to perform a different analysis on the same data, can you?
What is Reproducible Neuroimaging?
Scientific reproducibility
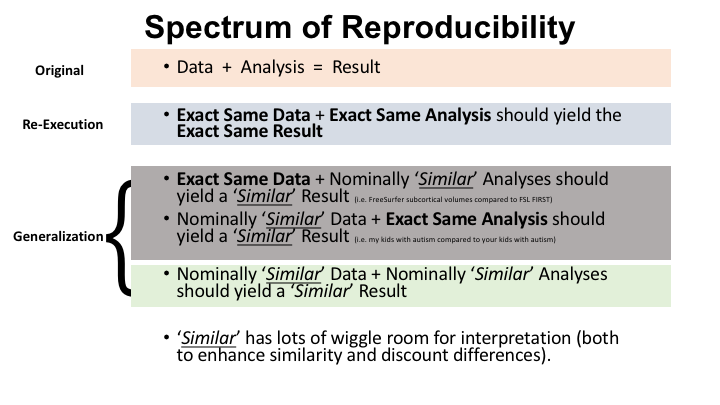
First, what do we mean when we say that scientific results are reproducible? Reproducibility is a broad term and is often used as a generic term to refer to replicate at some level the findings of a study. Here, we use the term scientific reproducibility in general to mean re-executability, that is, the ability to obtain the exact same results when the same data is analyzed using the exact same analysis methods. Re-executability focuses on methodological reproducibility and should be seen as simply the first step in establishing the validity of a given set of results. A result, that is, a claim made about the meaning of a study, is fully reproducible when it can be re-executed and generalized independently. Some of what we are referring to as generalization is referred to as replication, defined as repeating an entire study with new data to see if the original results can be replicated. But as you can see in the graphic below, there are multiple types of replication in the age of open data and tool sharing. A true biological inference should hold true regardless of the methods used to observe a biological process or the specific sample of the population being studied.
Reproducible Neuroimaging
Building on the materials above, reproducible neuroimaging refers to the practice of conducting and disseminating neuroimaging research in a manner that allows others to independently verify and replicate the findings. It encompasses a set of principles, practices, and tools aimed at ensuring transparency, rigor, and accountability in neuroimaging studies. Transparency is achieved through others being able to know precisely ‘what operations’ were performed on ‘what data’ . Remember, the “others” that benefit from reproducible neuroimaging include:
- You
- Future you
- Your lab
- Your colleagues
- Reviewers
- Other neuroimagers
- Data scientists
- AI
What does reproducible neuroimaging look like in practice?
ReproNim’s principles of reproducible neuroimaging
Study planning:
- Implement good science basics, e.g., power analysis, statistical consult
- Consider using pre-existing data for planning and/or analysis
- Create an NIH-compliant data management and sharing plan
- Adopt open consent to allow broad sharing of data
- Pre-register your study
Data and metadata management:
- Use standard data formats and extend them to meet your needs.
- Use version control from start to finish
- Annotate data using standard, reproducible procedures
Software management:
- Use released versions of open source software tools.
- Use version control from start to finish
- Automate the installation of your code and its dependencies
- Automate the execution of your data analysis
- Annotate your code and workflows using standard, reproducible procedures
- Use containers where reasonable
Publishing everything: publishing re-executable publications
- Plans should be shared (pre-registration)
- Software should be shared
- Data should be shared
- All research objects should be FAIR
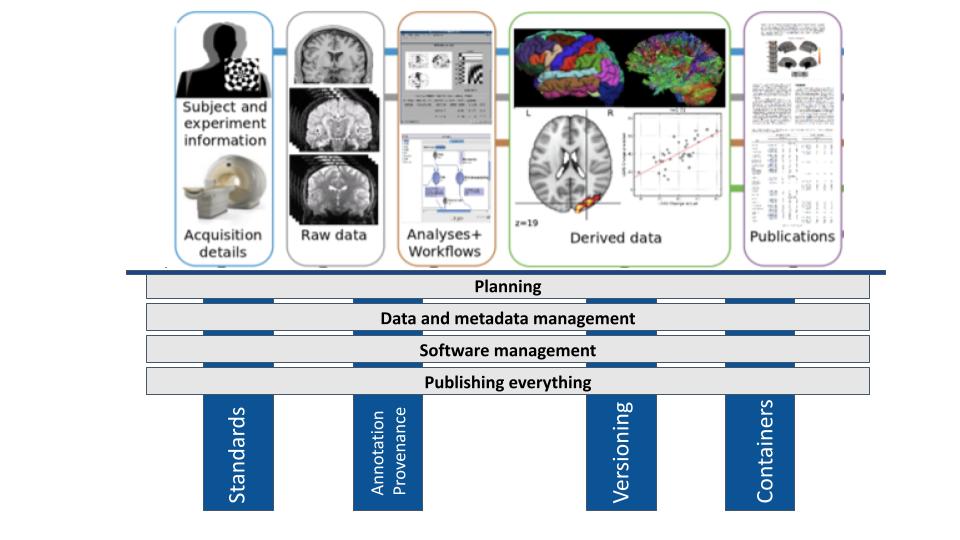
In turn, as indicated by the blue highlights in the above figure, four core actions are key to implementing these principles:
Use of Standards: Using standard data formats and extending them to meet specific research needs is important for data and metadata management in reproducible neuroimaging.
Annotation and provenance: Annotating data using standard, reproducible procedures ensures clarity and transparency in data management (data and metadata management). Provenance refers to the origin and history of data and processes, enabling researchers to track how data was generated, modified, and analyzed (data and metadata management, software management, and publishing everything). This is essential for understanding the context of data and ensuring reproducibility.
Implementing version control: Version control is crucial for both data and software management. It allows researchers to track changes over time, revert to previous versions if necessary, and collaborate effectively.
For data, version control helps manage different versions of datasets and track modifications made during processing and analysis (data and metadata management).
For software, version control helps track code changes, manage different versions of analysis scripts, and ensure that the correct version of the code is used for each analysis (software management).
And even publications can be versioned (publishing everything).
Use of Containers: Containers provide a portable and self-contained environment for running software, ensuring that the analysis can be executed consistently across different computing environments (software management). They encapsulate all the software dependencies needed to run an analysis, making it easier to share software (publishing everything) and reproduce results.
How does ReproNIM help support reproducible neuroimaging?
ReproNim’s goal is to improve the reproducibility of neuroimaging science, while making the process of capturing and precisely describing all essential/necessary experimental details both easier and more efficient for investigators. ReproNIM focuses on practices and tools that support researchers and software engineers in integrating reproducible principles and actions into their own neuroimaging workflow. We call this operational framework for reproducible analysis the ReproSystem.
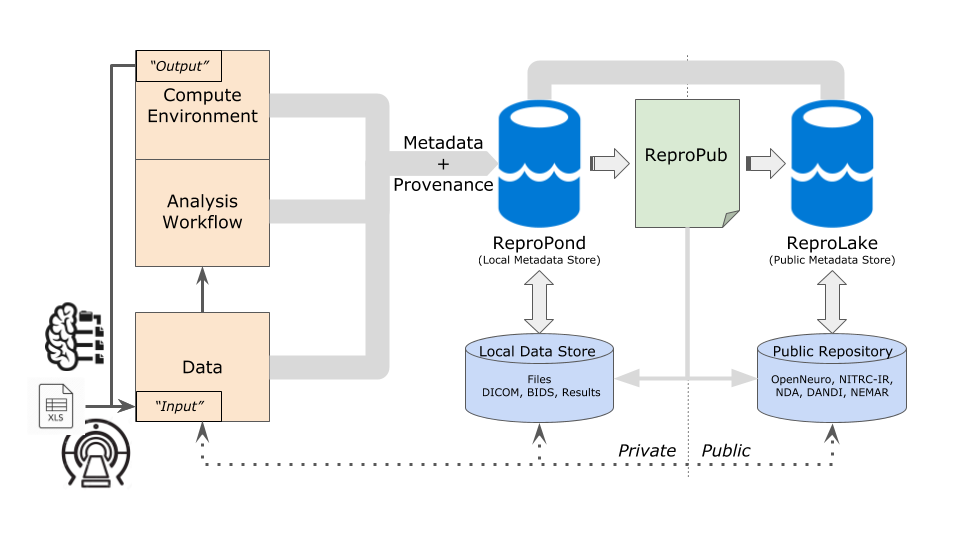
As outlined in the accompanying figure, the ReproSystem framework takes the full experimental cycle of neuroimaging data acquisition, analyses, and publication into account. This system explicitly includes both data (any of various types, such as scanner acquisition, data converted to BIDS, behavioral data, output of prior analysis) entering into an analysis, and the metadata+provenance (i.e. software versions and computer operating system versions used, processing script, subject demographics, results description, etc.) of the analysis. The metadata maintains a link to its associated data permitting flexibility regarding how each of these types of information sources can be used. To the familiar components of data and analysis environments, we also add to the framework a means to store and retrieve machine-readable description of metadata associated with the tools in both a local/private store (which we refer to as a ReproPond) and a shared/public repository (ReproLake).
To get started, see:
- Getting started with ReproNim
- The ReproGuide to tools and use cases